Abstract
Hepatocellular Carcinoma (HCC), a major subset of liver
cancer, stands as a global health concern, accounting for over
700,000 annual deaths annually. Current treatments offer limited
prognostic efficacy, leading to persistent complications and tumor
recurrence. A deeper comprehension of novel targets against HCC
progression is imperative due to ineffective clinical treatments.
Currently, many cell transduction pathways have been identified and
correlated to excess Collagen I production in HCC, as Collagen I is
an indicator of HCC progression; however, the involvement of
Transforming growth factor-β1 (TGF-β1), which plays an important
role in HCC development, and the gene encoding Collagen type I
(COL1A1) in HCC remains unclear. This study aims to elucidate the
previously unknown relationship of the TGFβ pathway and the COL1A1
gene in HCC by testing how TGF-β1 regulates the expression level of
COL1A1 and its impact on the proliferation and migration of
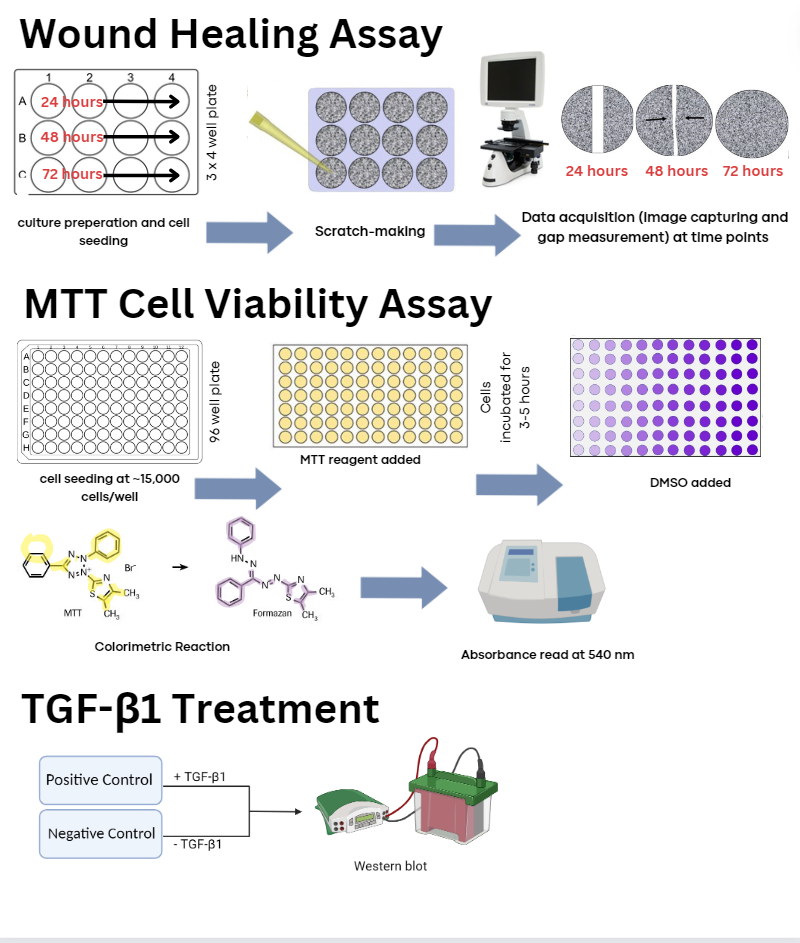
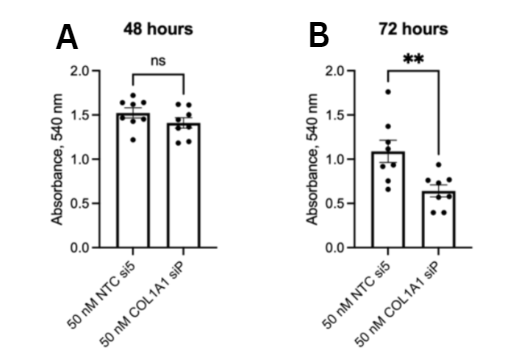
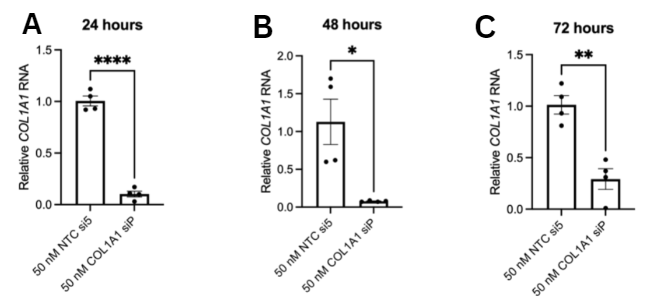
hepatocellular carcinoma cells. HepG2 cells were cultured and COL1A1
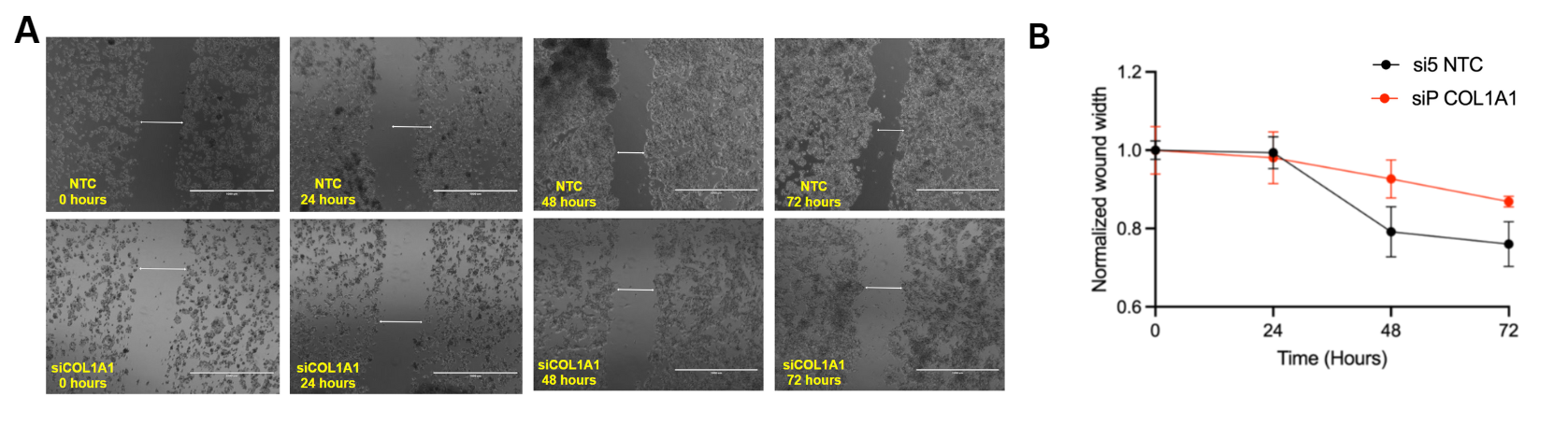
was knocked down via siRNA transfection to assess changes in
proliferation and migration rates of HCC, in vitro. These cells were
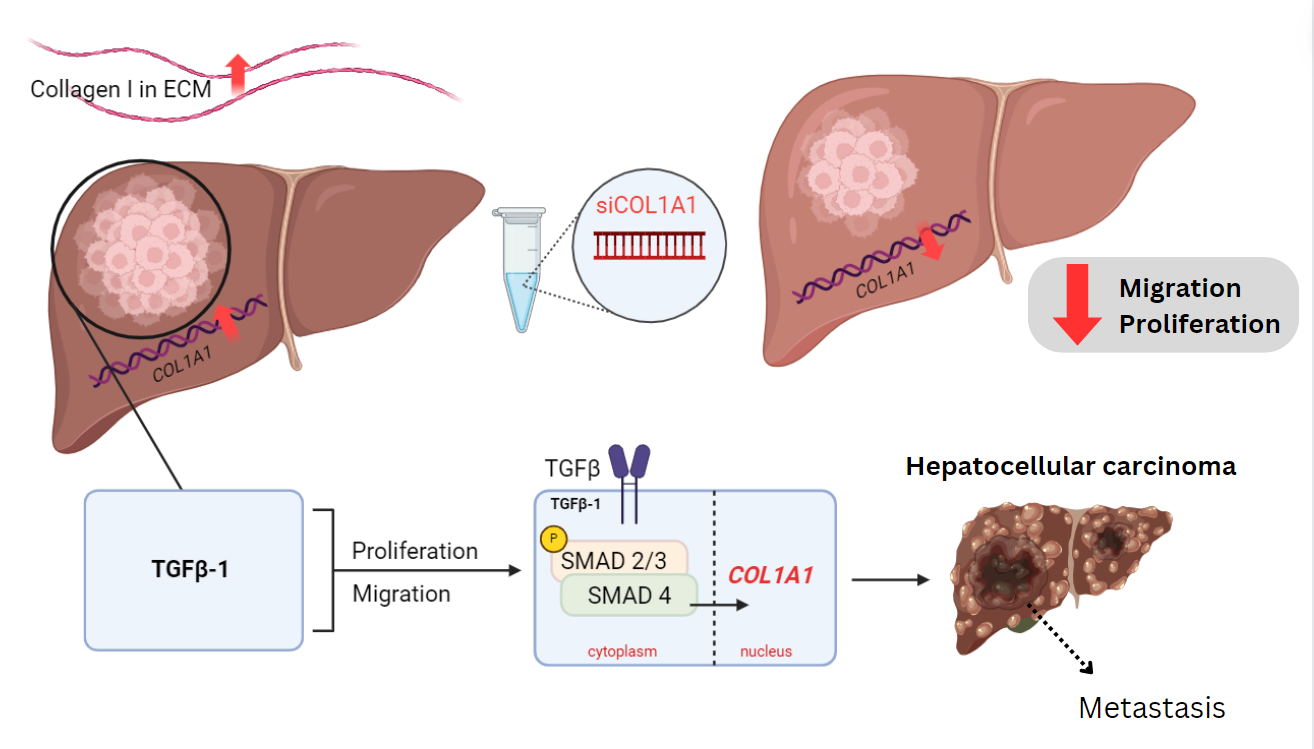
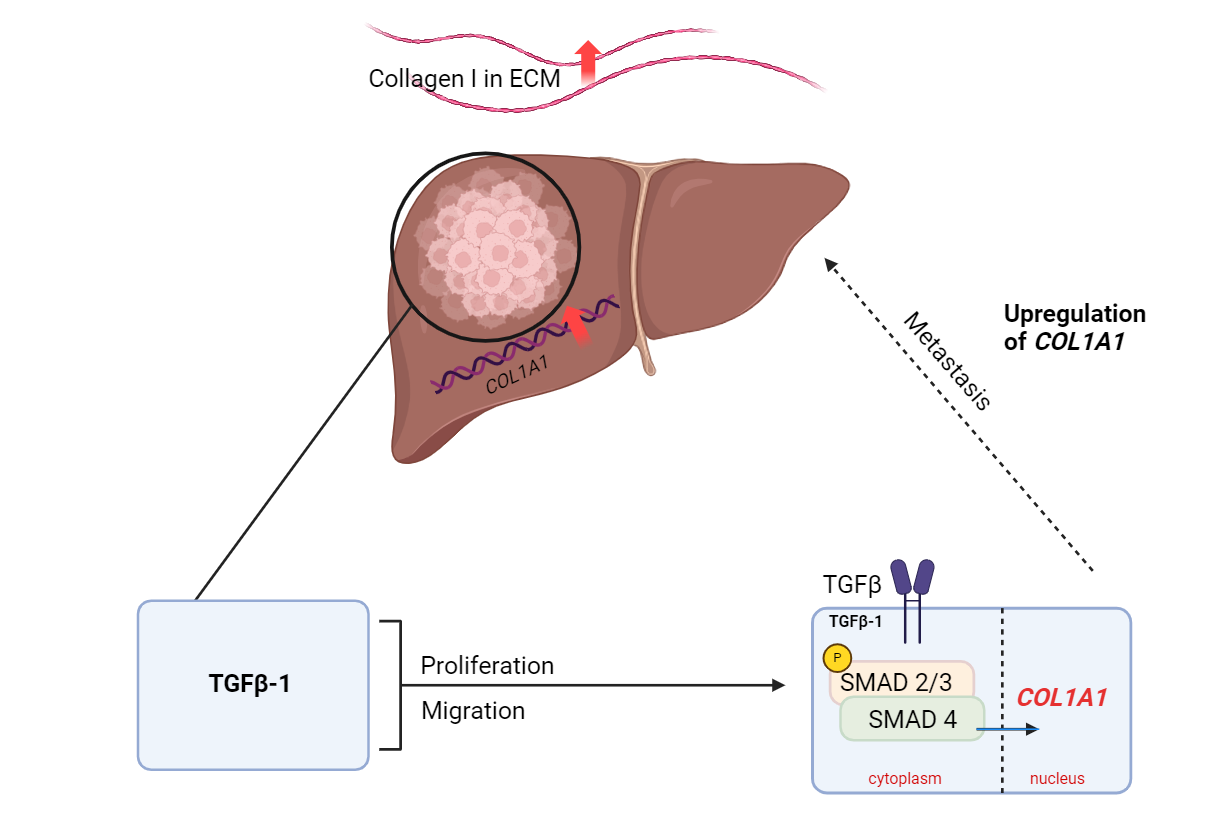
then treated with TGF-β1. Based on the findings, COL1A1 is
overexpressed in HCC. Upregulation of COL1A1 facilitates the
proliferation and migration of HCC cells through the TGFβ pathway.
The correlation of TGF-β1 and COL1A1 upregulation introduces a novel
therapeutic target against HCC, deterring HCC cell progression by
reducing ambient levels of collagen I in the carcinoma tissue.
The graphical abstract was created in BioRender (BioRender, n.d.).