Background
Sleep is an important part of life, taking up about a third of a person’s lifetime. Yet, there are many sleep disorders
that can affect an individual’s ability to sleep or their experiences while sleeping. Narcolepsy, a rare and chronic
neurological disorder that negatively impacts the control of sleep-wake cycles, manifests in different and sometimes
even dangerous ways throughout the day in patients. Narcoleptic patients experience excessive daytime sleepiness (EDS),
even if they felt rested after waking up. This leads to the possibility of falling asleep in the middle of an activity,
such as talking. In more dangerous cases, this can happen when a person is driving, leading to fatal car accidents
(de Mello et al., 2013). When this sudden onset of sleep occurs, the person experiences rapid eye movement sleep (REM);
in this phase of sleep, they are unable to move. Another potentially life-threatening symptom of narcolepsy is
cataplexy. The two types of narcolepsy, Type 1 (NT1) and Type 2 (NT2), are differentiated by the presence of cataplexy
as a symptom in those with NT1. Cataplexy involves sudden weakness in the muscles, rendering them limp or unable to
move. Often triggered by strong emotions such as laughter, cataplexy can range from a mild period of weakness in a
few muscles to a complete collapse of the body (NINDS, 2023).
Due to the rarity of the disease, affecting about one in 2,000 people, the pathophysiology of narcolepsy has yet to be
fully explored. Narcolepsy with cataplexy is characterized by the deficiency of hypocretin, also known as orexin–a
neuropeptide that plays a role in stimulating wakefulness. The loss of hypocretin-producing neurons in the hypothalamus
of the brain contribute to their deficiency in narcoleptic patients (Liblau et al., 2015). Furthermore, symptoms of
narcolepsy, particularly cataplexy, can look similar to epileptic seizures (Diukova et al., 2021). As narcolepsy
appears to have common symptoms with epilepsy, misdiagnosis is not rare, especially in the case of children and
adolescents due to the onset of narcolepsy (Baiardi et al., 2015). Misdiagnosis presents a significant diagnosis
challenge in clinical practice, and electrophysiological studies are necessary for differential diagnosis (Baiardi et
al., 2015). An electroencephalogram (EEG), frequently used in the diagnosis of sleep and neurological disorders,
records the electrical signals from the brain. The Multiple Sleep Latency Test (MSLT) checks EDS by measuring the time
it takes for someone to go from the start of a daytime nap to the first signs of sleep, known as sleep latency. In
addition, the Epworth Sleepiness Scale (ESS) is a questionnaire that is reliably used in studies to measure the
frequency of daytime sleepiness (Zhang & Zhao, 2008). In the case of narcoleptic patients who are incorrectly
diagnosed, it will take substantially more time to acquire proper diagnosis and treatment, if at all.
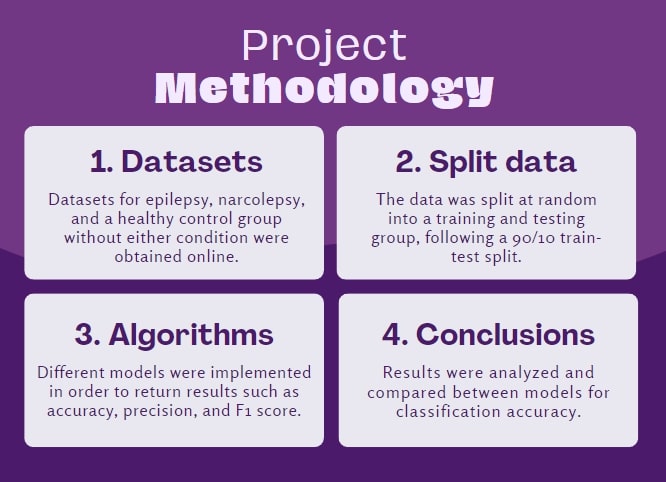
Recently, many machine learning algorithms have been trained to help identify different disorders from EEG recordings.
As EEGs are an important diagnosis tool for both sleep disorders and neurological conditions, there is an abundance of
EEG data for both narcolepsy and epilepsy patients. Machine learning classification models are trained with datasets of
both healthy and affected individuals; as these models become increasingly accurate, their current clinical
implications play a growing role in diagnosis of diseases. When using EEG recordings for image classification, one of
the most commonly used algorithms is a convolutional neural network (CNN) (Pawan & Dhiman, 2023). CNNs are useful for
working with two-dimensional data, and are composed of many layers–the highest-level building blocks in deep learning.
Each layer will receive input from a previous layer, and after transforming the values it obtains, it then passes these
values to the next layer. The convolutional layers of a CNN perform calculations at each part of the image, which helps
it identify specific details. Afterwards, the pooling layers take this data and simplify it. Activation functions in
the output layer such as the Rectified Linear Unit (ReLU) and sigmoid are then used in learning relationships between
features. Ultimately, the model is able to extract key features and improve its accuracy on predictions through more
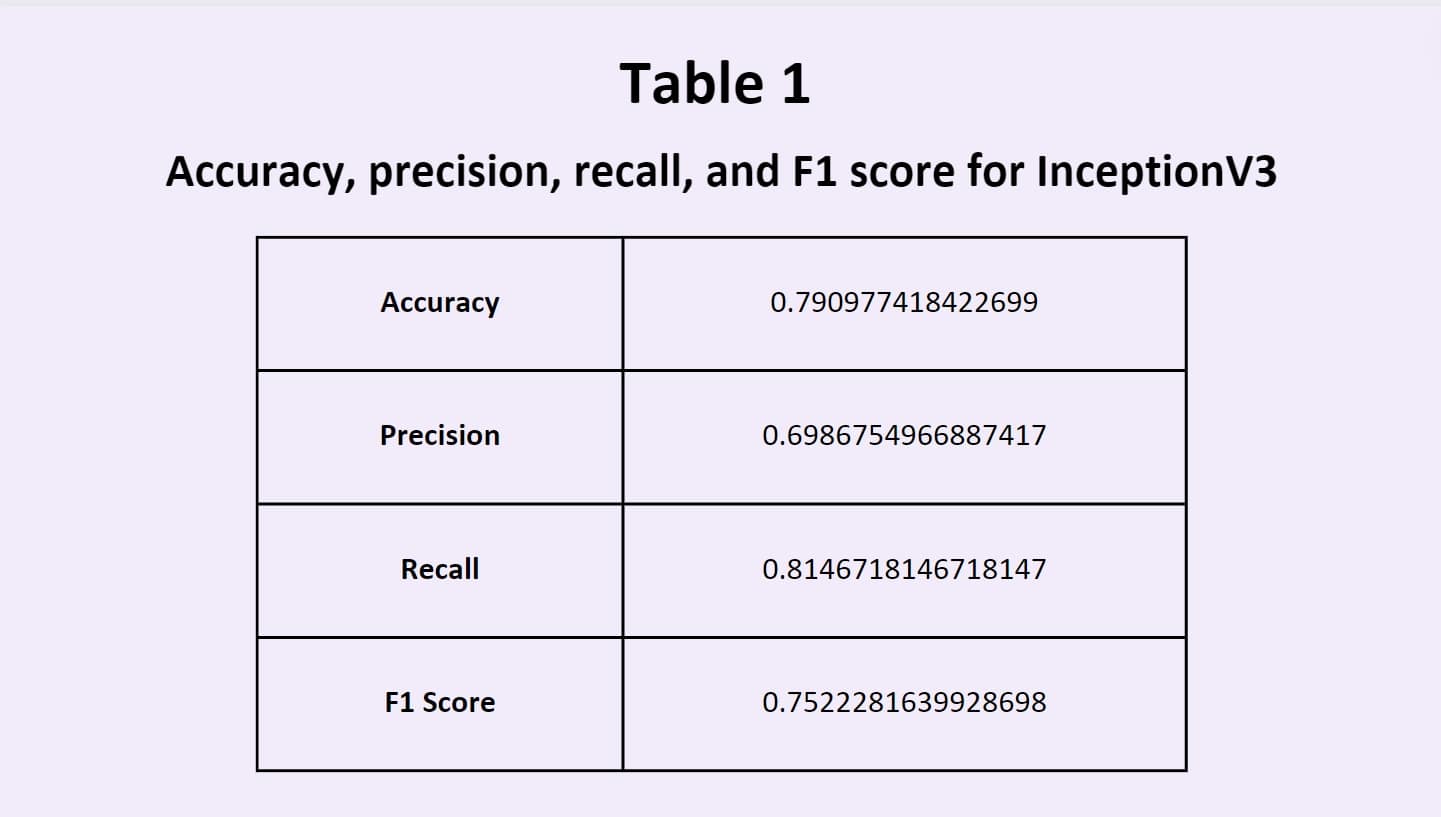
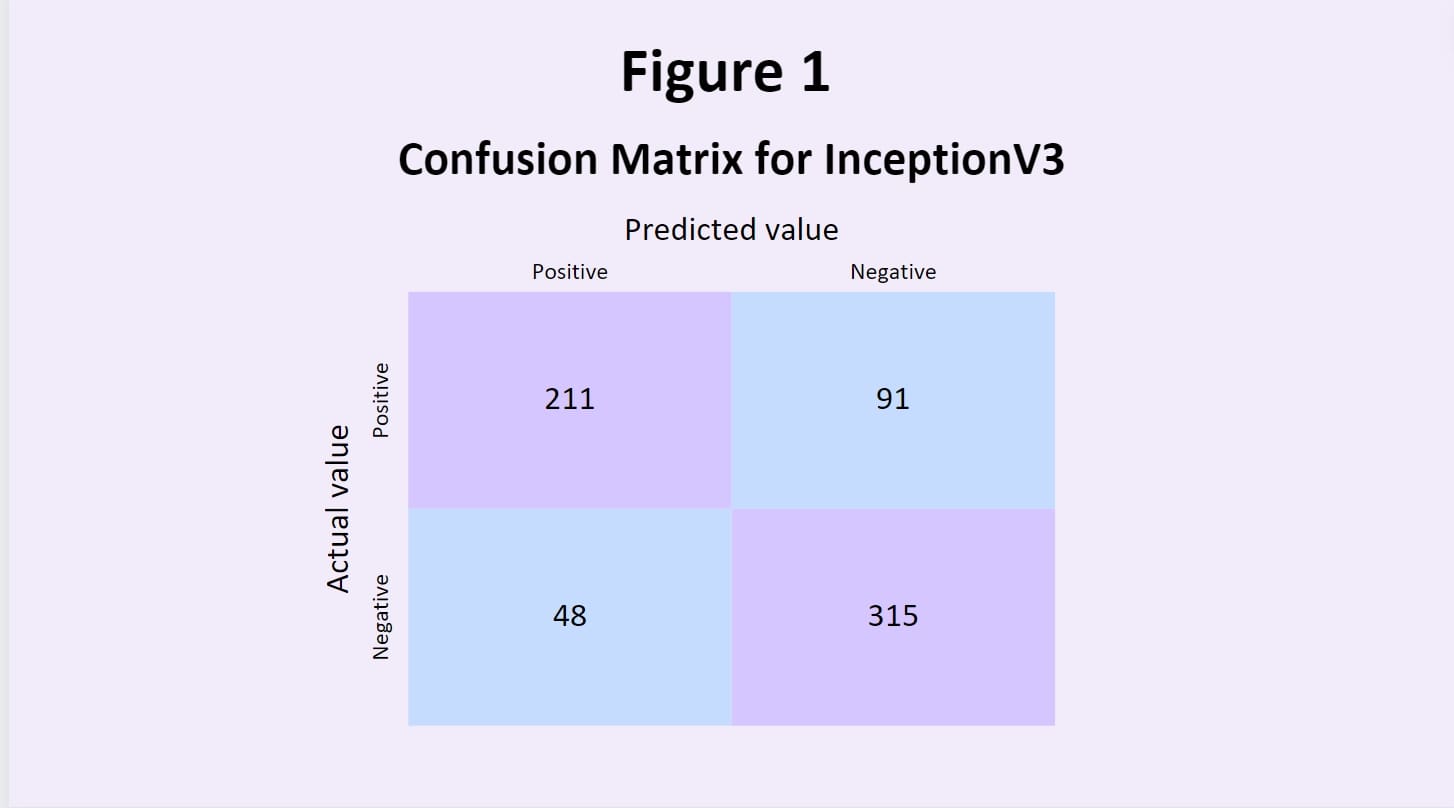
training data. One of the most common models used is Inception V3, a type of CNN; it is 48 layers deep and offers both
good accuracy and computational cost, making it a robust model for image classification. In addition, ensemble models
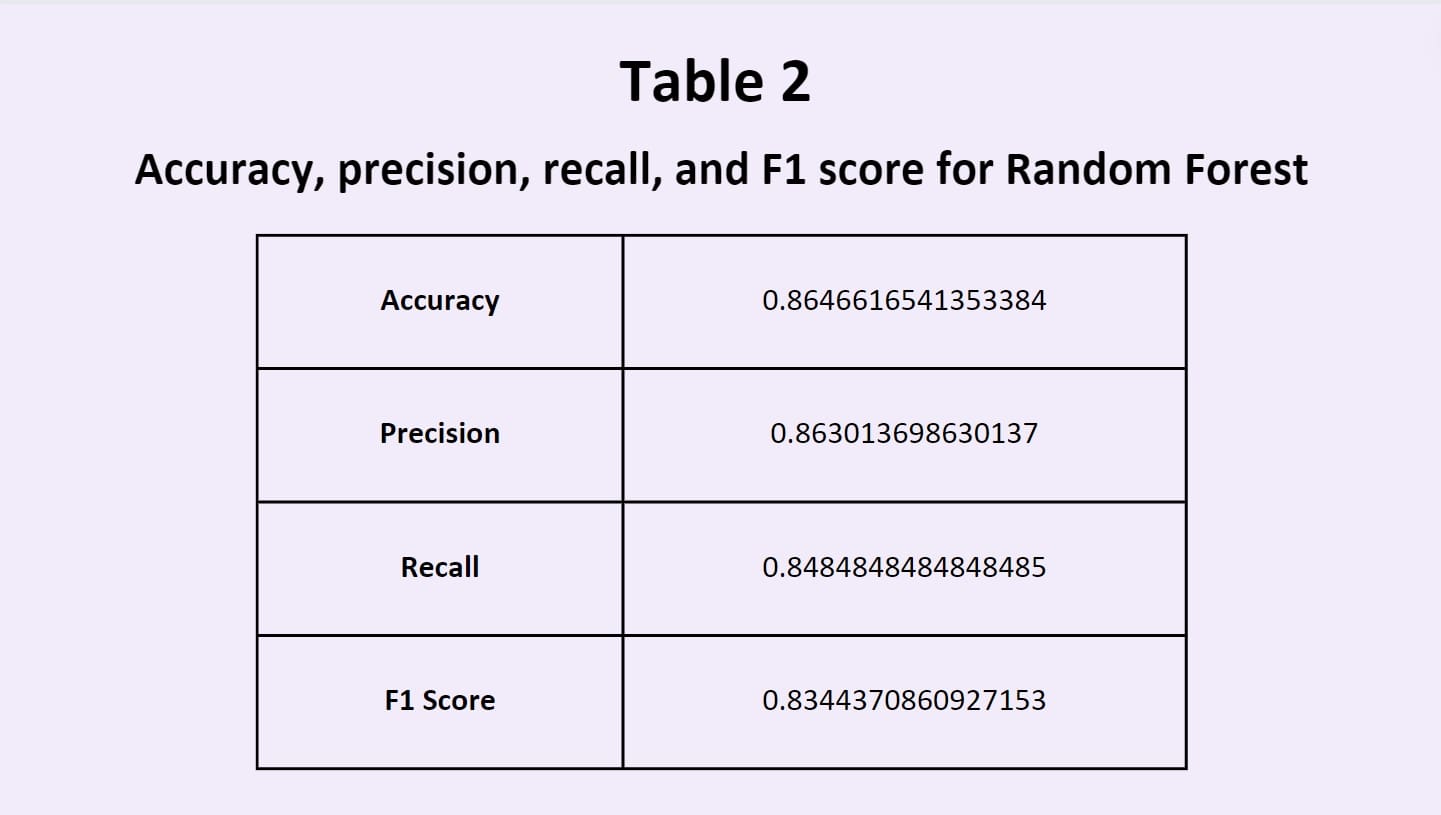
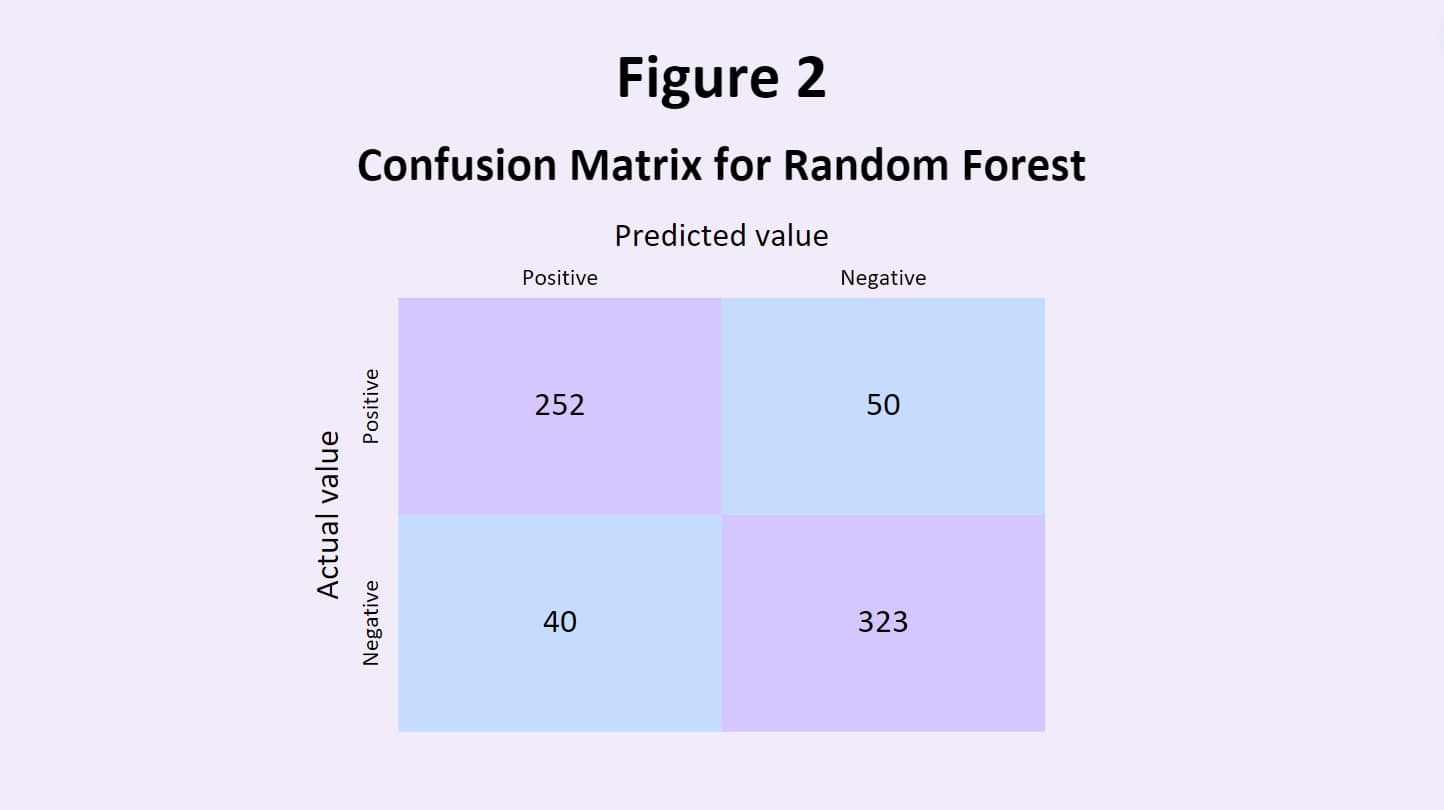
such as random forests utilize the predictions of many different models and merge them together to evaluate their own
classification system. This aims to even out the biases of individual models, and leverages the collective decisions
and intelligence of all the models that it draws from (Mahajan et al., 2023).