Developing a Predictive Model to Compute the Progression of Alzheimer’s Disease via a Neural Apoptotic Marker
Rishab Nair
Advisor: Kevin Crowthers, Ph.D.
Overview
The overall aim of this project is to optimize the diagnostic and therapeutic processes of patients with Alzheimer’s Disease by developing a predictive model to detect and predict the prognosis of the disease from earlier stages.
Abstract
This research project aimed to assess the validity of apoptotic marker behavior in predicting the trajectory of Alzheimer's disease, given its increasing prevalence and global health impact. The study utilized fMRI datasets to examine the behavior of specific apoptotic markers in individuals with Alzheimer's, focusing on Cytochrome C, Caspase-3, -7, and -9. Due to time constraints, resting state fMRI data from rats with Alzheimer's-like pathology was used as a substitute. A predictive model was developed, analyzing pixel-by-pixel representations of fMRI scans over two to 21 weeks. The model achieved a 95% accuracy in predicting Alzheimer's progression at 21 weeks, demonstrating the potential of apoptotic marker behavior for early disease detection and personalized treatment planning. This discovery not only sheds light on Alzheimer's complexities but also suggests potential applications for understanding and managing other neurodegenerative diseases involving neuronal apoptosis.
Graphical Abstract
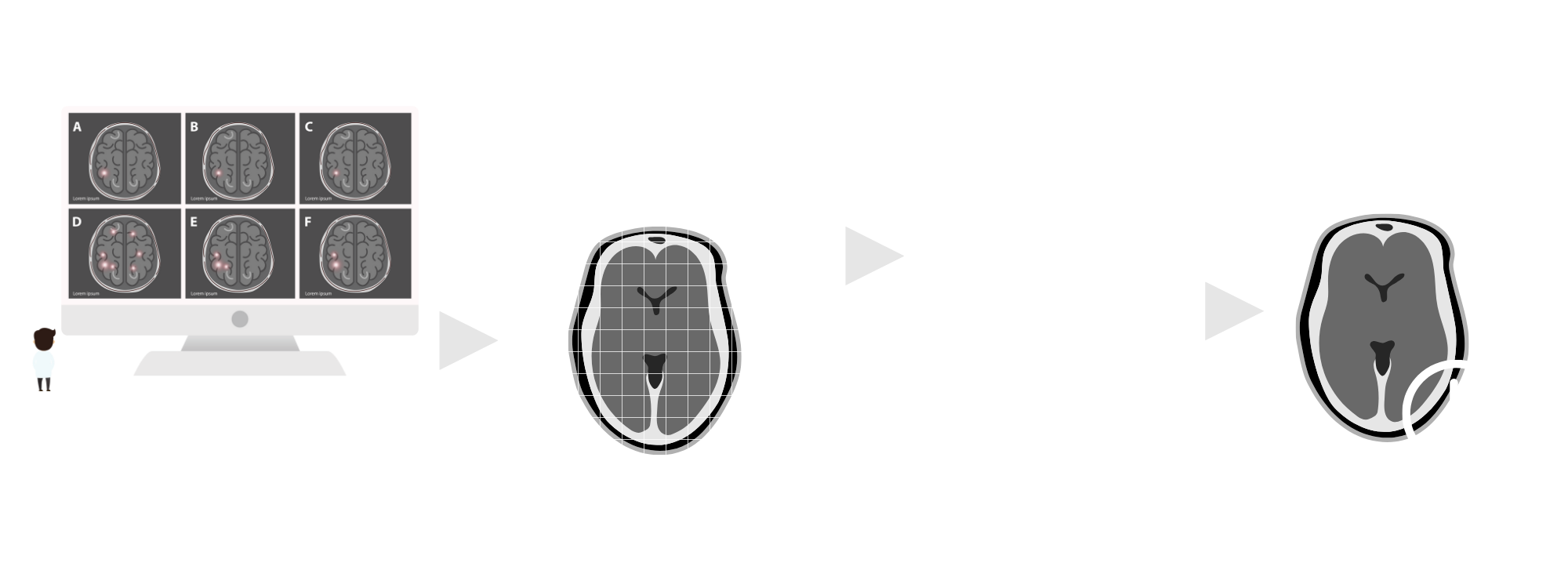
Problem Statement
Current methods of Alzheimer’s diagnosis occur late into the progression of the disease, and detriment the effectiveness of therapeutic programs. Current therapeutic plans are relatively generalized as well, although multiple different versions of the disease occur.
Objective
This project seeks to mitigate these issues by creating a predictive model aiming for efficiency in detecting the disease in earlier stages, as well as calculation of its most-likely progression by using the rate of activity of apoptotic markers found within the brain.
Background Infographic
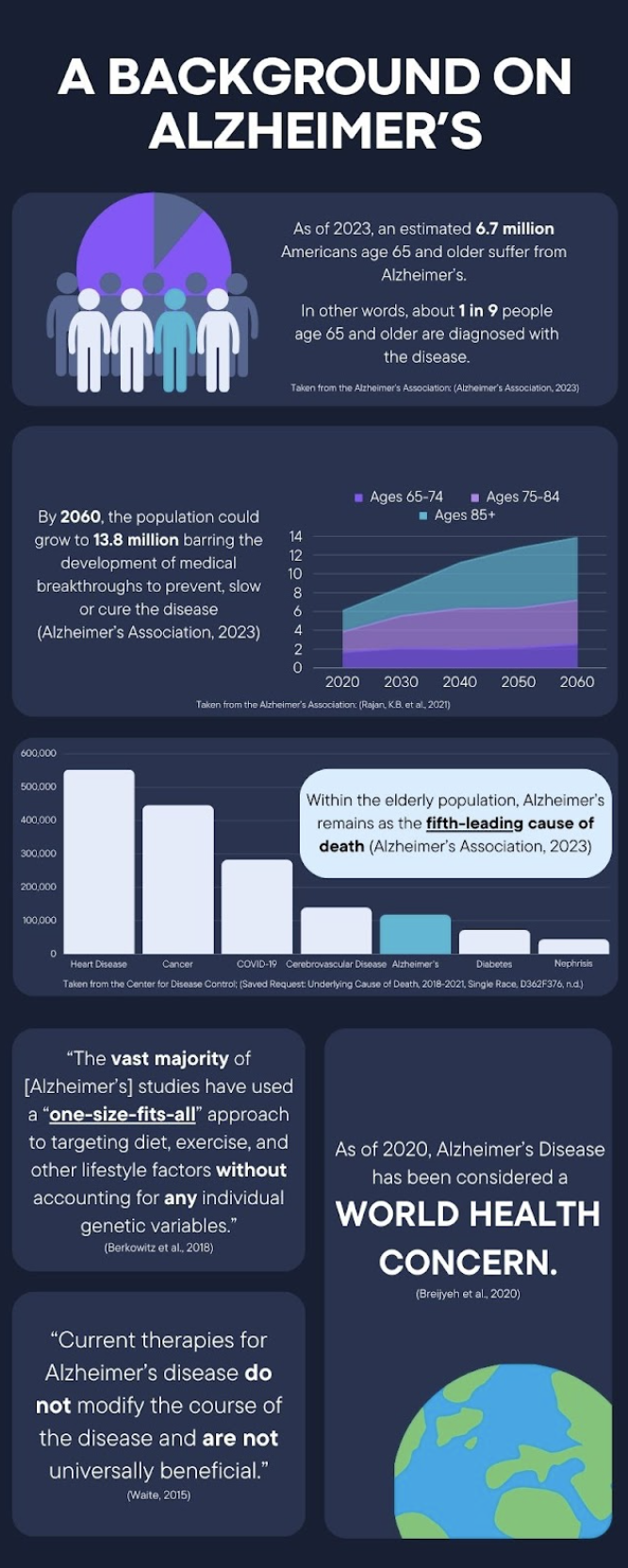
Background
As of 2023, an estimated 6.7 million Americans age 65 and older suffer from Alzheimer’s. Within this population, Alzheimer’s remains as the fifth-leading cause of death. By 2060, the population could grow to 13.8 million barring the development of medical breakthroughs to prevent, slow or cure the disease (Alzheimer’s Association, 2023). Alzheimer’s does not solely have such significance in just America, however; as of 2020, the disease has been classified as a world health concern (Breijyeh et al. 2020).
This project consists of two phases: determining a marker that can detect areas in the brain affected by Alzheimer’s disease and using this marker to develop a predictive model to determine the progression of the disease so that medical professionals obtain a greater understanding of its behavior.
It is known that massive neuronal death in Alzheimer’s disease is a result of apoptosis, or programmed cell death (Shimohama, 2000). Past studies on cell death and apoptosis have focused on potential markers that can detect the causation of apoptosis. However, these studies tend to center on general cells– not neurons. In its first phase, this project aims to close this research gap by determining a possible correlation of neuronal cells to existing cell death marker studies. The project aims to determine a specific cell death marker to detect affected areas in Alzheimer’s.
Current methods of Alzheimer’s diagnosis occur late into the progression of the disease, and detriment the effectiveness of therapeutic programs (Emory, 2011). Current therapeutic plans are also relatively generalized, although multiple different versions of the disease occur. Therefore, in its second phase, this project seeks to mitigate these issues by creating a predictive model. This model aims for efficiency in detecting the disease in earlier stages, as well as prediction of its progression. Using this information, doctors can optimize and personalize the patients’ therapeutic plans.
Procedure Infographic

Procedure
The successful execution of this project necessitates the utilization of diverse datasets, ranging from classic Alzheimer's MRI brain scans to specialized brain scans designed for monitoring Caspases -3, -7, and -9. These datasets serve as crucial inputs for the development and training of the model, enabling it to recognize patterns and correlations relevant to the targeted objectives. Additionally, the implementation of Python libraries, particularly SciKit-Learn, is integral to the project's computational framework. SciKit-Learn, a powerful machine learning library, facilitates tasks such as data preprocessing, feature selection, and model evaluation. Its incorporation adds a robust and efficient dimension to the project, enhancing the overall analytical capabilities and contributing to the successful deployment of the predictive model.
Thorough background research was conducted using easily accessible sources to understand possible markers associated with apoptosis within Alzheimer's disease, such as Cytochrome C, executioner caspases -3, -7, or both. After preliminary research, Functional Magnetic Resonance Imaging (fMRI) scans, the most common type of brain scan used for targeted imagery, was then used to determine the markers' significance in identifying the damaged brain regions via their activation. The next stage was gathering various brain scan datasets–from public online datasets–to examine and validate patterns from the potential markers. The procedure reiterated the fMRI step in the case where the pattern for one potential marker didn’t hold strength.
To gauge the path of Alzheimer's disease, a predictive model was finely tuned to highlight the identified marker patterns. Initially, a basic model was implemented, then consistently refined to boost accuracy in prediction.
However, before delving into the complexities of constructing a predictive model, it is imperative to adopt a more foundational approach. The initial step involved the creation of a classification model. This fundamental model served as a precursor, enabling the identification and categorization of data based on predefined classes or labels. By establishing this basic framework, the groundwork was laid for more advanced predictive modeling. Classification models provide a structured starting point, aiding in the systematic analysis of data patterns and paving the way for the development of more sophisticated predictive algorithms. The created model used basic MRI scans, refitting them to 100x100 grayscale images, and employed the K-Nearest Neighbors algorithm.
To achieve 90% accuracy, the model was adjusted from classification to prediction and fine-tuned by fitting it to the specialized brain scans designed for monitoring Caspases -3, -7, and -9. This process entails multiple iterations where different predictive models are generated and meticulously evaluated. Various machine learning algorithms, parameter configurations, and feature engineering techniques are explored to identify the optimal combination that maximizes accuracy. Rigorous testing against diverse datasets and cross-validation methods was employed to ensure the generalizability of the model. This iterative approach, driven by continuous experimentation and analysis, aims to push the predictive model beyond the 90% accuracy threshold, thereby enhancing its reliability and efficacy in predicting the trajectory of Alzheimer's disease.
Regrettably, the intended fMRI scans for monitoring caspase activation were not secured within the designated timeframe. To address this setback, a proof of concept was devised using fMRI scans of rats treated with streptozotocin, a chemical meant to mimic the effects of Alzheimer’s. This strategic shift aimed to showcase the feasibility of constructing a predictive model using fMRI scans, despite the unavailability of the originally planned data on caspase activation.
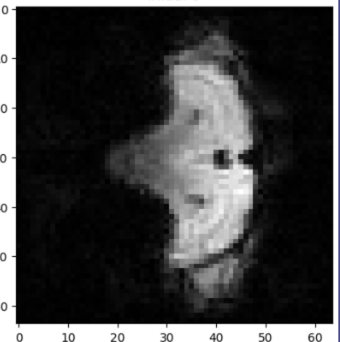
Figure 1: An example of a visualization of a slice of an fMRI scan of a rat treated with streptozotocin, through pixel-by-pixel color value metrics.
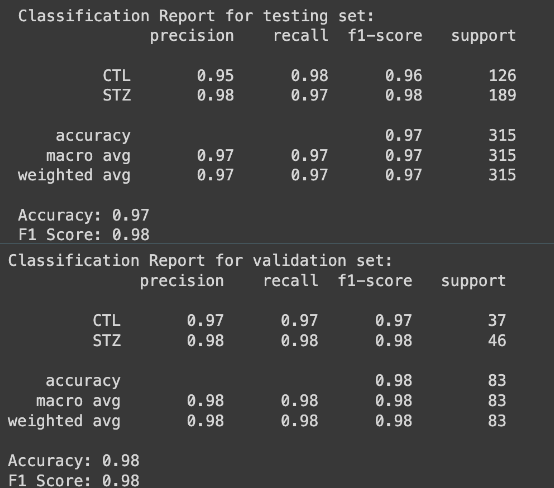
Figure 2: Classification reports of the testing process (top) and the validation confirmation (bottom)
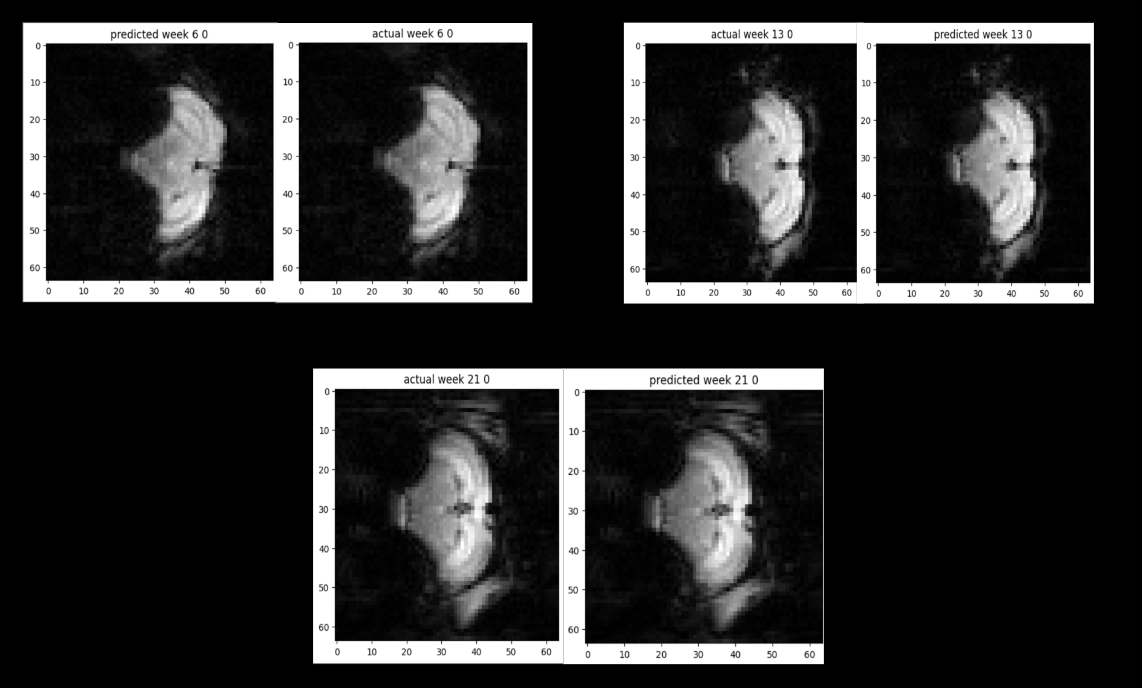
Figure 3: Actual fMRI scans compared to the model’s prediction over the course of 6(top left), 13(top right), and 21(bottom) weeks.
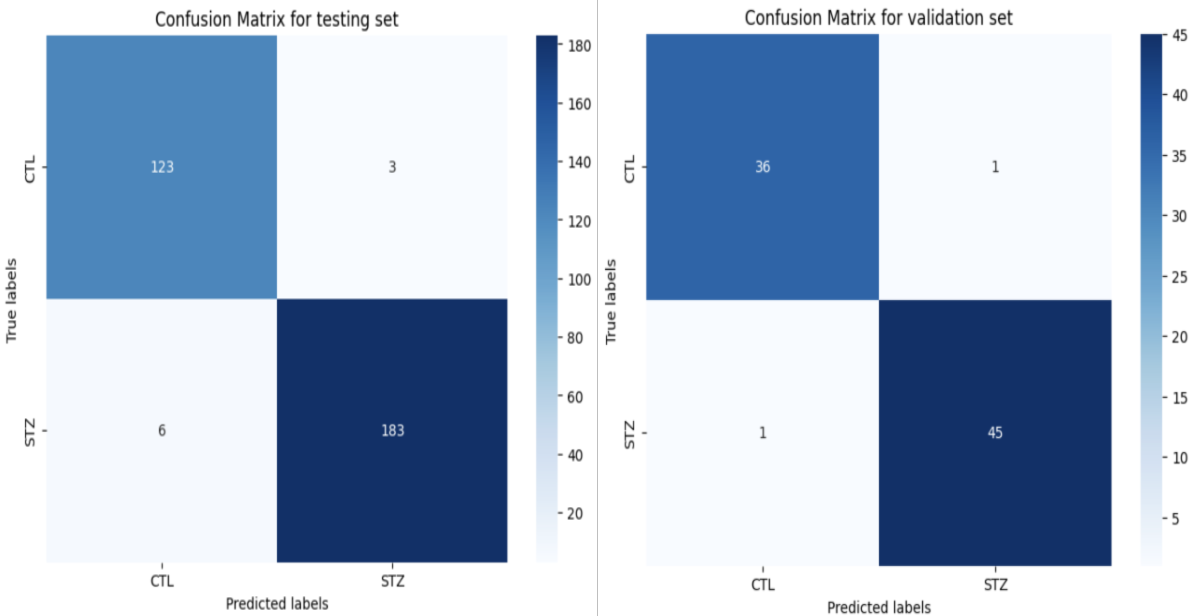
Figure 4: Confusion matrices of the testing process (left) and the validation confirmation (right)
Analysis
The developed model utilizes pixel-by-pixel analysis to classify and predict future fMRI scans. The classification accuracy of the model, using K-Nearest Neighbors stands at an impressive 98%, indicating its proficiency in accurately categorizing fMRI data. To validate against overfitting, the model underwent testing against a separate set, achieving a validation accuracy of 97%. Moreover, the model demonstrates robust performance with a statistical F1-score of 95%, affirming its precision and reliability in identifying patterns within fMRI scans. Temporal Consistency and Prediction Accuracy In addition to classification, the model's predictive capabilities were evaluated by matching pixel-by-pixel across different time settings. This approach facilitates the understanding of temporal changes within fMRI scans. The prediction accuracy of the model remains high at 95%, underscoring its effectiveness in forecasting future scans while maintaining fidelity to the intricate dynamics of brain activity over time.
Discussion
The main objective of the project was to demonstrate the feasibility of using functional Magnetic Resonance Imaging (fMRI) to detect and monitor the progression of streptozotocin in rats. Although the project did not achieve its ultimate goal due to time constraints, it successfully established a strong proof of concept. The data analysis revealed compelling evidence supporting the hypothesis. fMRI data from both control and streptozotocin-treated rats were analyzed to identify distinct patterns indicative of streptozotocin presence and progression. Through statistical analysis, the accuracy of the model in detecting treated rats was calculated, achieving approximately 98% accuracy in the testing set and 97% accuracy in the validation set. This robust performance indicates the reliability and effectiveness of the approach. To assess the reliability of the model, the F1-score, a widely used metric in artificial intelligence for evaluating classification performance, was employed. With F1-scores averaging around 95% for both classification and progression tasks, the model demonstrated high reliability in accurately identifying and monitoring streptozotocin-induced changes in rats' brain activity.
Throughout the project, various challenges and limitations required careful consideration and mitigation strategies. Initially, distinguishing between control and treated rats proved challenging due to subtle differences in fMRI data. However, by implementing z-score normalization techniques, these differences were enhanced, and the objective was achieved. Additionally, potential confounding variables such as variations in rat physiology and imaging artifacts were addressed through meticulous experimental design and data preprocessing steps. Standardizing experimental procedures and employing rigorous quality control measures minimized the impact of these variables on the results, ensuring the reliability and validity of the findings. While the project did not fully accomplish its overarching objective within the allotted timeframe, it provided valuable insights and laid the foundation for future research in this domain. Lessons learned from this experience, including the importance of robust data normalization techniques and comprehensive validation procedures, will inform future iterations of the project and contribute to its eventual success.
In conclusion, despite not achieving all predefined objectives, the project succeeded in establishing a solid proof of concept for using fMRI to detect and monitor streptozotocin-induced changes in rats. The high accuracy and reliability of the model underscore its potential utility in preclinical research and hold promise for advancing the understanding of neurochemical processes.
This work represents a significant advancement in the application of predictive modeling within clinical bioinformatics, offering the potential for diagnoses with increased certainty (Pais, 2022). The focus on Alzheimer’s is particularly noteworthy, as it addresses the pressing need to aid the approximately 6 million individuals currently suffering from the disease (Alzheimer’s Association, 2023).
Future research will focus on expanding the established proof of concept by integrating specific apoptotic markers into fMRI scans of rats, aiming to extend these findings to human scans. This progression seeks to deepen our understanding of the neurochemical changes associated with apoptosis across preclinical and clinical settings. One crucial aspect of future research involves the selection of appropriate apoptotic markers. Identifying markers with high specificity and sensitivity to neuronal apoptosis is essential to ensure clear and reliable signals detectable through fMRI imaging techniques. Additionally, refining experimental protocols will be necessary. This includes optimizing imaging parameters, standardizing procedures, and implementing rigorous quality control measures to enhance the detection of apoptotic markers while minimizing confounding variables and artifacts. Advanced data analysis techniques, such as machine learning algorithms and pattern recognition methods, will play a crucial role in interpreting complex fMRI data. These methods will help extract meaningful information related to apoptotic processes from the collected data. Translating findings from animal models to human studies will require careful validation and refinement of imaging protocols. Longitudinal studies conducted in clinical populations and healthy controls will provide valuable insights into the role of apoptosis in various neurological disorders. Exploring the clinical implications of detecting apoptotic markers using fMRI will be another critical avenue of future research. This includes investigating the potential utility of these markers as diagnostic biomarkers, prognostic indicators, and therapeutic targets for neurodegenerative diseases and other neurological conditions. By advancing research in this direction, future studies can contribute to a deeper understanding of the molecular mechanisms underlying neurodegeneration and facilitate the development of novel diagnostic and therapeutic strategies aimed at mitigating neuronal loss and preserving brain function.
Conclusion
In this study, the primary objective was to demonstrate the feasibility of utilizing functional Magnetic Resonance Imaging (fMRI) to detect and monitor streptozotocin-induced changes in rats. The methodology involved fMRI scans of both control and streptozotocin-treated rats, followed by rigorous data analysis techniques. The results revealed promising findings, with distinct patterns of brain activity observed in streptozotocin-treated rats compared to controls. Through statistical analysis, high accuracy was achieved in detecting treated rats, indicating the reliability of the approach. Additionally, the utilization of the F1-score demonstrated the robustness of the model in accurately identifying and monitoring streptozotocin-induced changes in brain activity. The analysis of relevant data highlighted the successful proof of concept for utilizing fMRI in detecting streptozotocin-induced alterations in rats, despite encountering initial challenges. This study lays a solid foundation for future research aimed at further elucidating the neurochemical processes underlying streptozotocin-induced neurodegeneration.
These findings underscore the potential of fMRI as a valuable tool in preclinical research for detecting and monitoring neurochemical changes associated with neurodegenerative diseases. This study not only advances our understanding of streptozotocin-induced effects but also paves the way for the development of novel diagnostic and therapeutic strategies to combat neurological disorders.
References
Alzheimer’s Association (2023). 2023 Alzheimer's disease facts and figures. Alzheimer's &
dementia : the journal of the Alzheimer's Association, 19(4), 1598–1695. https://doi.org/10.1002/alz.13016
Bhardwaj, A., Kishore, S., & Pandey, D. K. (2022). Artificial Intelligence in Biological Sciences. Life
(Basel, Switzerland), 12(9), 1430. https://doi.org/10.3390/
Breijyeh, Z., & Karaman, R. (2020). Comprehensive Review on Alzheimer's Disease: Causes and
Treatment. Molecules (Basel, Switzerland), 25(24), 5789. https://doi.org/10.3390/
McIlwain, D. R., Berger, T., & Mak, T. W. (2013). Caspase functions in cell death and disease.
Cold Spring Harbor perspectives in biology, 5(4), a008656. https://doi.org/10.1101/
Naniche, N., Sau, D., & Pasinelli, P.. (2011). In Vivo and In Vitro Determination of Cell Death
Markers in Neurons. Methods in Molecular Biology, 793, 9–21. https://doi.org/10.1007/
Nano, M., Mondo, J. A., Harwood, J., Balasanyan, V., & Montell, D. J. (2023). Cell survival
following direct executioner-caspase activation. Proceedings of the National Academy of Sciences, 120(4), e2216531120. https://doi.org/10.1073/
Pais, R. J. (2022). Predictive Modelling in Clinical Bioinformatics: Key Concepts for
Startups. Biotech (Basel (Switzerland)), 11(3), 35. https://doi.org/10.3390/
Shimohama, S. (2000). Apoptosis in Alzheimer's disease--an update. Apoptosis : an international
journal on programmed cell death, 5(1), 9–16. https://doi.org/10.1023/
Stadelmann, C., Deckwerth, T. L., Srinivasan, A., Bancher, C., Brück, W., Jellinger, K., &
Lassmann, H. (1999). Activation of caspase-3 in single neurons and autophagic granules of granulovacuolar degeneration in Alzheimer's disease. Evidence for apoptotic cell death. The American journal of pathology, 155(5), 1459–1466. https://doi.org/10.1016/
Waite, L. M. (2015). Treatment for Alzheimer's disease: has anything changed?. Australian
prescriber, 38(2), 60–63. https://doi.org/10.18773/
Poster
PDF Viewer unavailable. Download PDF Here