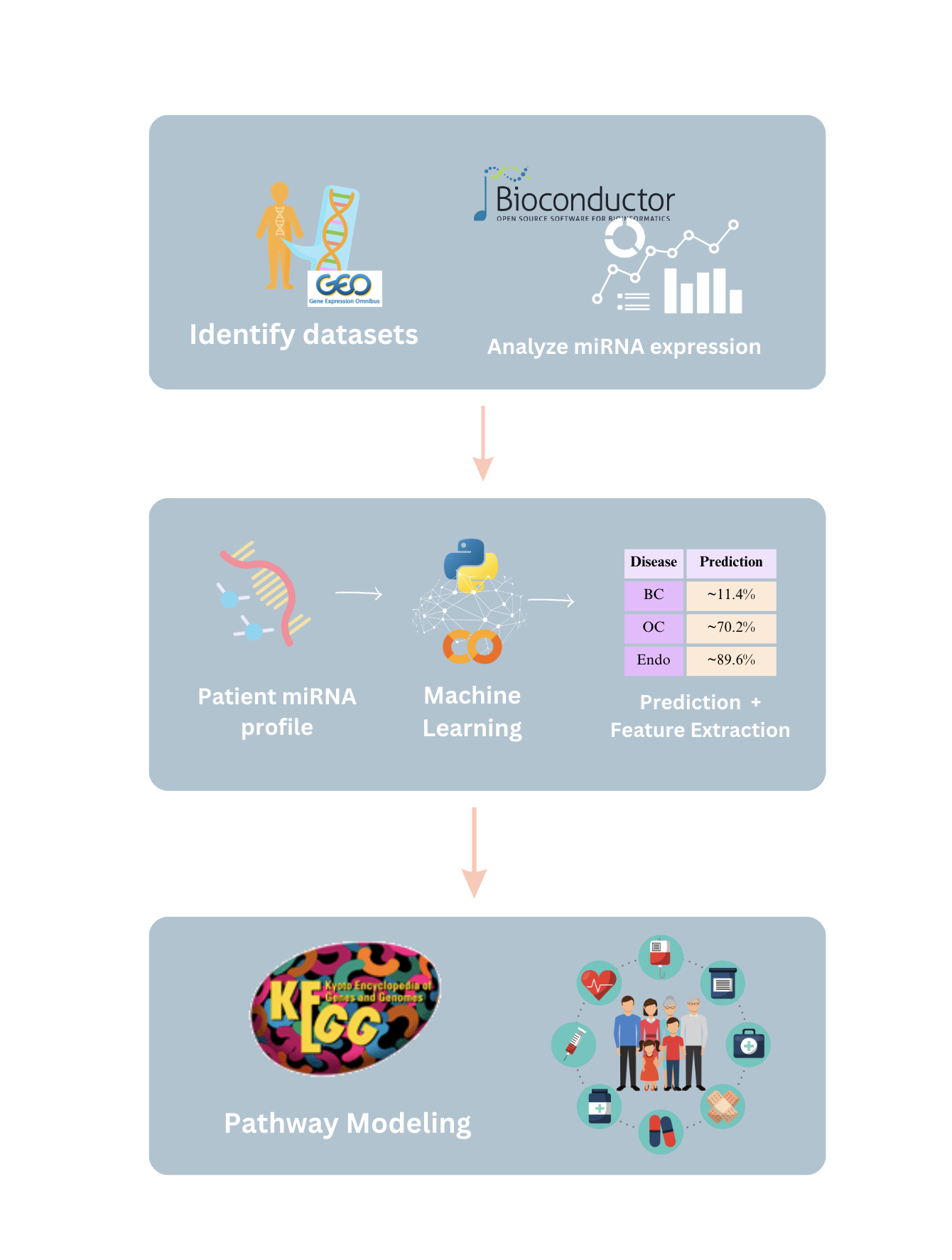
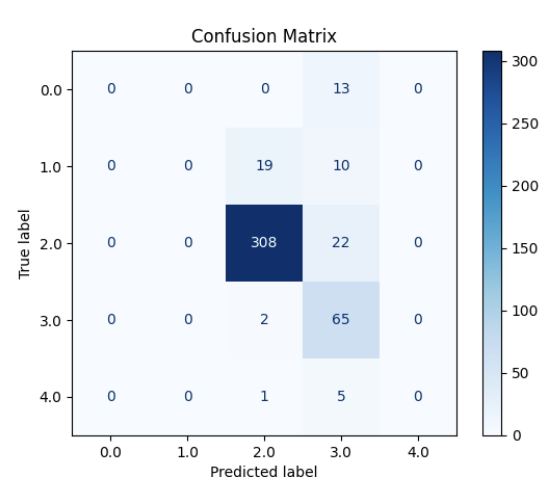
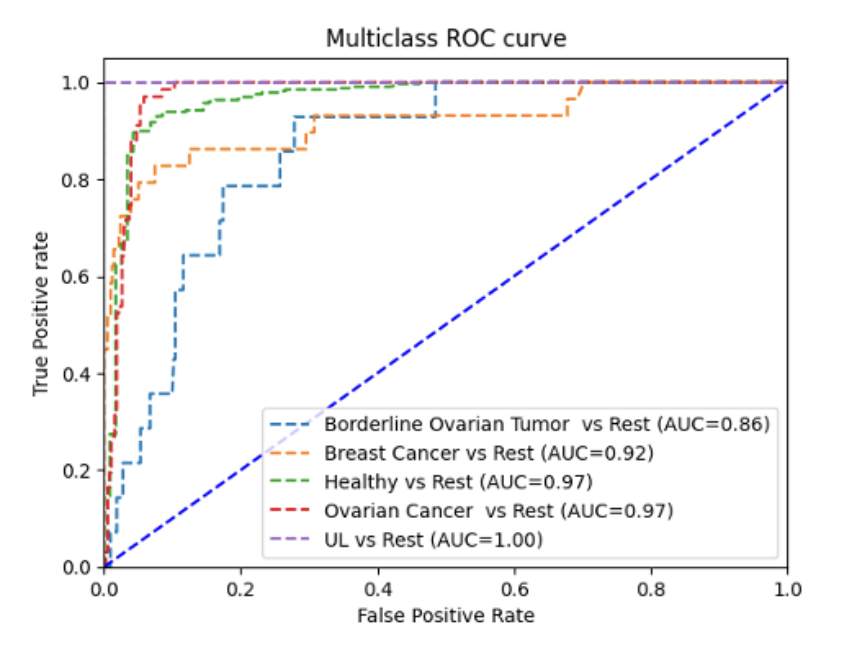
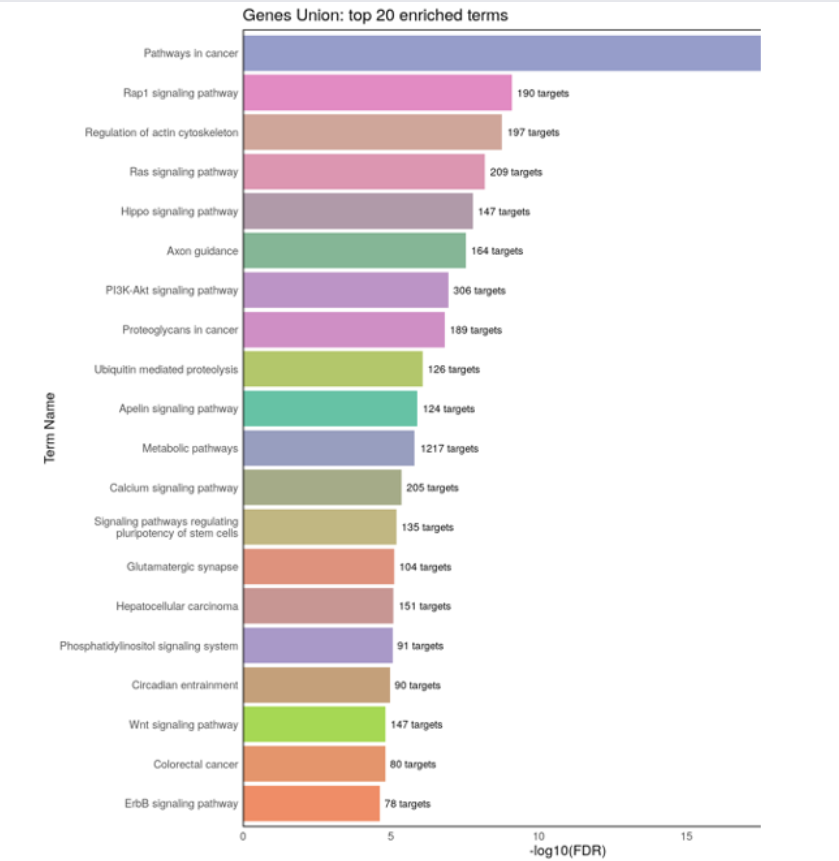
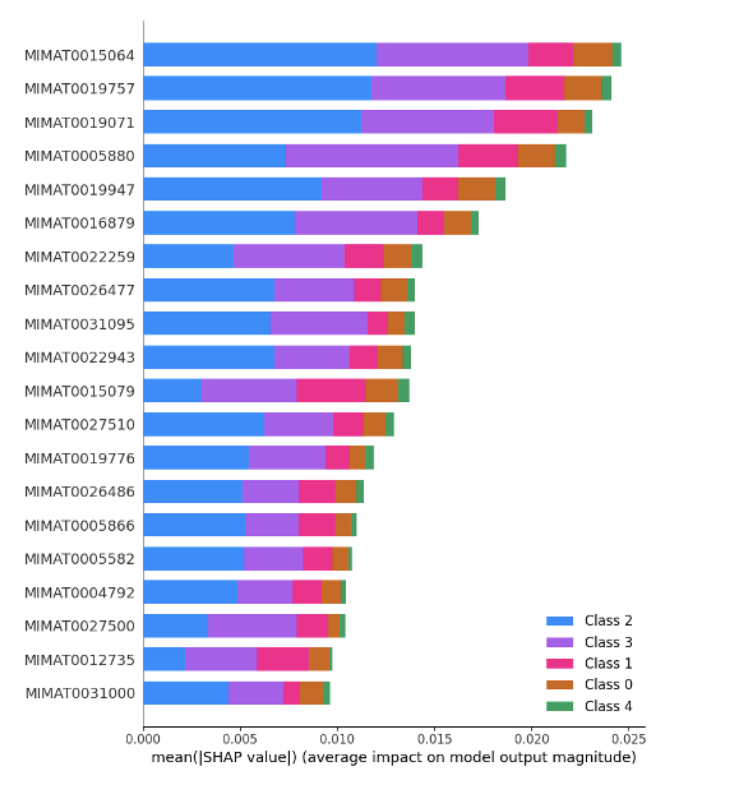
Women’s health faces a growing crisis: 1 out of 10 women will experience chronic gynecological diseases, yet treatment can be delayed up to 4-11 years due to the social stigma, ambiguous symptoms, complex pathology, lack of noninvasive and accessible diagnostic tools, and cost-ineffective drug discovery pipelines. MicroRNAs (non-coding RNA segments) offer promise as noninvasive biomarkers as they regulate gene expression and are easily detectable in bodily fluids. This study aims to create a deep-learning pipeline that utilizes blood-based miRNA expression to facilitate early disease detection and identify therapeutic targets. Logistic Regression, Random Forest, Deep Neural Network, and Ensemble models were trained on more than 2000 publicly accessible patient miRNA samples to differentiate diseases, specifically ovarian cancer, breast cancer, endometriosis, and polycystic ovary syndrome. The models performed greater than 90% accuracy for binary classification and 85% for multi-disease classification. Feature extraction techniques, such as Shapley analysis, were applied to identify key biomarkers and perform pathway analysis and gene clustering to understand the unique and shared pathology behind malignant and cancerous female conditions. Notably, miR-let-7d emerged as a consistently dysregulated miRNA, impacting the RAS signaling pathway, while miR-1307-3p has been linked to chemoresistance in ovarian cancer and endometriosis malignancy. These distinguished miRNAs provide insights into early disease detection biomarkers and potential therapeutic targets. This pipeline can be applied in a clinical setting by extracting miRNAs from patient blood samples and ev Overall, this pipeline aims to improve health outcomes for female patients by creating avenues for accessible and noninvasive serum-based diagnostics, assessing treatment options, expediting personalized drug discovery pipelines, and reducing misdiagnosis and waiting periods.