stem 1
STEM and Science/Technical Writing, taught by Dr. Crowthers, allows students to pursue a five-month long individual research project on any topic of their choosing. Students learn valuable skills in long-term project management, reading scientific articles, and writing grant proposals and theses.
Inhibition of Gliadin-alpha2 Using Novel Peptide Synthesis
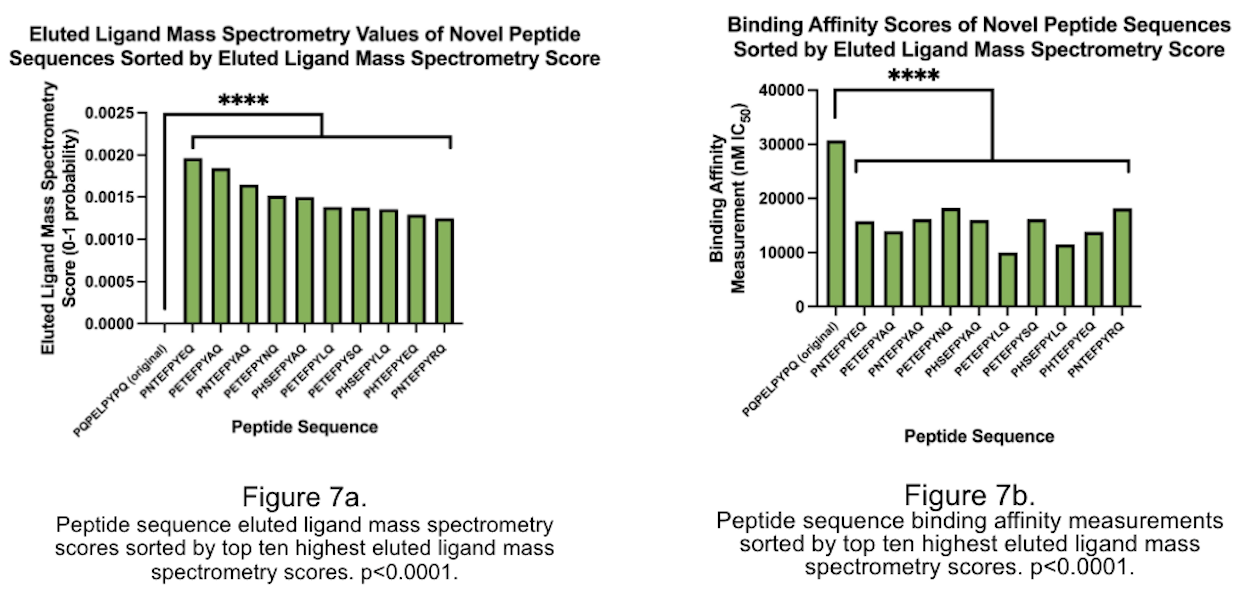
This project utilized NetMHCIIpan-4.1 to predict binding affinity and likelihood between peptide sequences and MHC-II molecules. The peptide sequence PETEFPYLQ is in the top 0.001 percent of both binding affinity and likelihood scores.
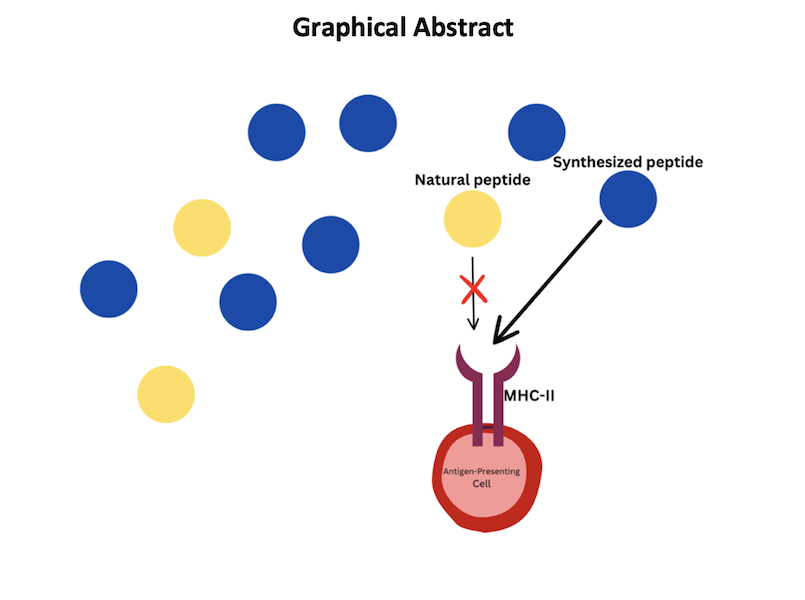
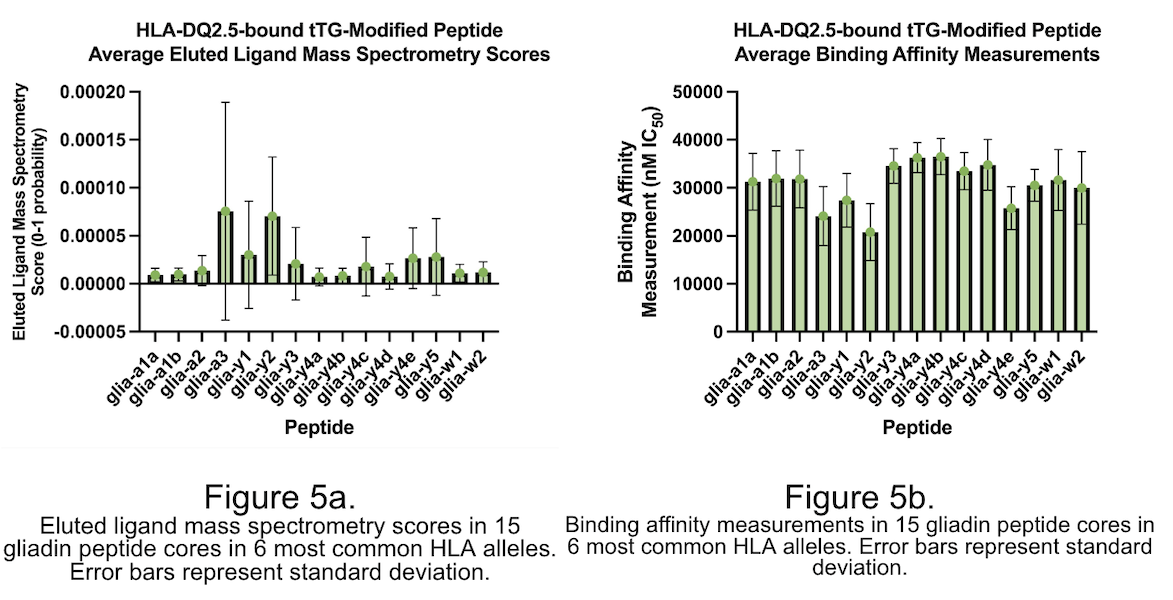
Affecting over 1 percent of the global population, celiac disease (CeD) is triggered upon the recognition of tissue transglutaminase (tTG)-modified peptide sequences by T-cells. Due to the possibility of gluten-free diets, little pharmacological research has been done into supplemental treatments other than enzyme-based therapies. This project utilized NetMHCIIpan-4.1 to predict binding affinity and likelihood between peptide sequences and MHC-II molecules. A peptide that competitively inhibits gliadin-alpha2 by binding to HLA-DQ2.5 was designed and tested in silico. The gene HLA-DQA0501-DQB0201 was the most susceptible to changes from tTG modification, getting altered by 200 percent. Gliadin-alpha3 and -gamma2 had the highest binding scores over the 6 most common HLA-DQ2.5 genes, sharing a glutamic acid in position 4. The peptide sequence PETEFPYLQ is in the top 0.001 percent of both binding affinity and likelihood scores. Specific peptide positions have an importance in binding scores and provided evidence for keeping specific amino acid positions the same as the original peptide sequence. This peptide treatment does not rely on the use of enzymes, and therefore opens the possibility to inhibit a larger range of peptides as they are not restricted to being degraded before entering the stomach and small intestine. Future work in this topic includes using the same methodology for a larger set of peptides.
Research Proposal
How do different sequences of amino acids impact the binding strength and binding probability of gliadin-alpha2 to the MHC-II molecule? Answering this question will result in the creation of a novel peptide that will competitively inhibit the binding of gliadin-alpha2 to the MHC-II complex.
There are two specific aims of this study: to determine which amino acid positions have the strongest effect on binding affinity and to determine a sequence of amino acids that will most effectively inhibit the tTG-modified gliadin-alpha2 peptide. This project uses NetMHCIIpan-4.1 to predict binding affinity and likelihood between the peptide and MHC-II molecule for most methodology.
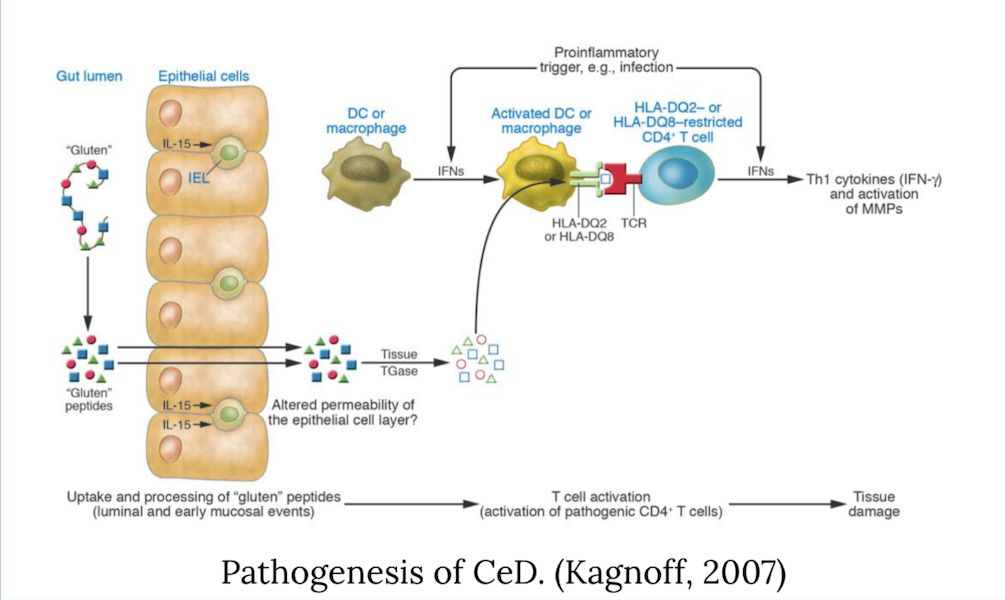
Celiac Disease (CeD) is a common food-induced inflammatory disease affecting 1% of the global population. It is triggered by ingesting wheat gluten or similar proteins found in cereals like barley and rye. The immune response attacks the small intestine, leading to damage on the villi, which promote nutrient absorption. Treatments include a gluten-free diet, but enzymatic treatments have been suggested for degrading gluten peptides. CeD relies on T-cell recognition (TCR) of gluten-derived peptides bound to HLA-DQ2 or HLA-DQ8 genes, which help the immune system identify harmful agents. Peptides, such as gliadins and glutenins, are non-degradable by most intestinal enzymes and are not degraded by most intestinal enzymes. The gliadin-𝛼2 peptide is of utmost importance in CeD patients as it has the most reported binding and cannot be naturally broken down. This peptide inhibits the immune system's reaction to the peptides, creating an autoimmune response damaging the patient's body. The goal of this project was determined to create a novel peptide that will competitively inhibit the binding of gliadin-𝛼2 to the MHC-II complex.
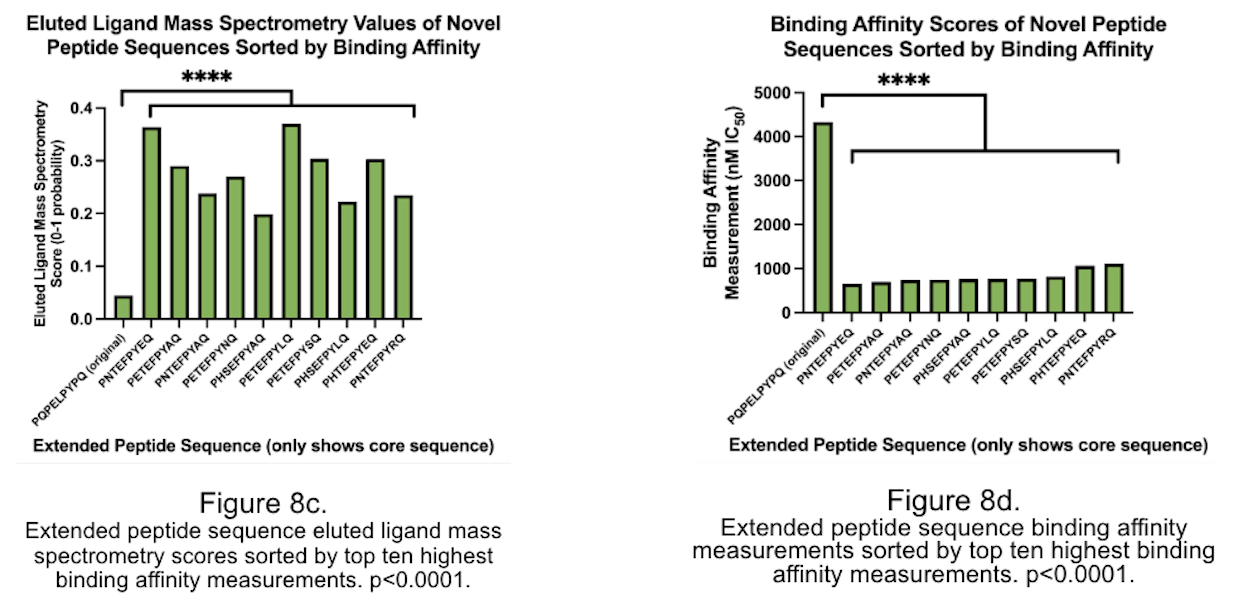
The project utilized web-based software and databases to analyze peptide sequences, binding affinity, and eluted ligand mass spectrometry scores. Amino acid sequences were based on gluten-derived proteins bound to the HLA-DQ2.5 allele, associated with CeD. The project used NetMHCIIpan-4.1 software from the Technical University of Denmark, Immune Epitope Database and Tools (IEDB) database from the National Institute of Allergy and Infectious Diseases, SYFPEITHI software from the University of Tübingen, and the RCSB Protein Data Bank (PDB) database from the Worldwide Protein Data Bank. The peptide core was modified by generating a list of permutation combinations representing amino acid positions that could be modified without changing the binding cleft. The top ten peptide sequences were re-sorted by highest to lowest score for eluted ligand mass spectrometry and lowest to highest binding affinity, respectively. A one-proportion Student's t-test was used to compare changes in binding affinity and eluted ligand mass spectrometry scores for modified and unmodified amino acid sequences of the same gluten-derived peptide.
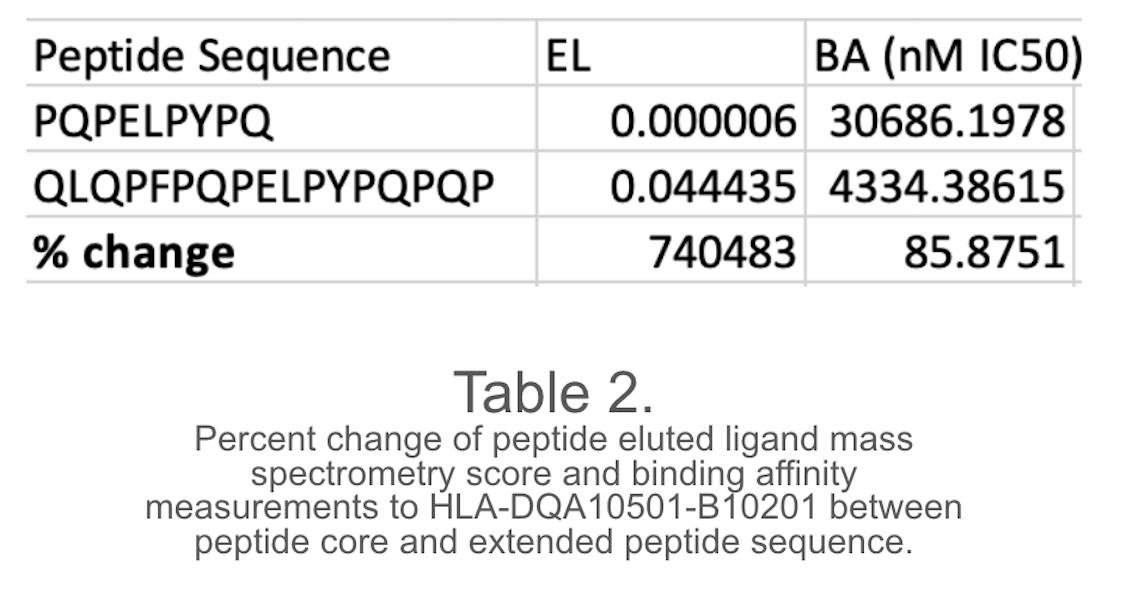
The study reveals that modifying peptide sequences, particularly position 4 to glutamic acid, can significantly alter binding affinity and eluted ligand mass spectrometry scores, often by over 50%. Furthermore, adding amino acids and extending the peptide sequence can alter these scores, with over 80% of BA scores altered and over 70% altered. The study also reveals that a majority of amino acids are more heavily influenced by changes in their codes. The aim is to develop novel peptides that can competitively inhibit gliadin-α2 if they maintain the same binding cleft. To achieve this, 20 sequences were created for highest binding affinity measurement and eluted ligand score. These sequences had statistically significant scores compared to the original BA and EL scores of the gliadin-a2 peptide, indicating they could effectively outcompete the natural gliadin peptide. The peptide sequence QLQPFPETEFPYLQPQP was found to be the most suitable for competitive inhibition of the gliadin-α2 peptide.
CeD is one of the most common diseases caused by eating food, affecting over 70,000,000 people, including over 3,000,000 Americans, where 60-70% of Americans who have the disease are undiagnosed and needlessly suffering (Celiac Disease Foundation, 2016). Little research has been done into CeD treatment because it has long been considered that a gluten-free diet is the only option. However, with newly developed biotechnologies and knowledge, it is now possible to develop a drug that targets the pathology itself. This project presents the possibility for a novel peptide-based approach using the sequence PETEFPYLQ by competitively inhibiting the gliadin-a2 peptide, the most prevalent gluten-derived peptide in CeD patients. This peptide could significantly reduce the effects of gliadin-a2 on the immune system and does not affect any CeD blood tests, proving it useful for CeD therapeutics.
Anderson, J. (2022). HLA-DQ2: The Primary Celiac Disease Gene. Verywell Health. https://www.verywellhealth.com/hla-dq2-the-primary-celiac-disease-gene-562569
Arentz‐Hansen, H., Körner, R., Øyvind Molberg, Hanne Quarsten, Vader, W., Kooy, Y., Knut E.A. Lundin, Koning, F., Roepstorff, P., Sollid, L. M., & McAdam, S. N. (2000). The Intestinal T Cell Response to α-Gliadin in Adult Celiac Disease Is Focused on a Single Deamidated Glutamine Targeted by Tissue Transglutaminase. Journal of Experimental Medicine, 191(4), 603–612. https://doi.org/10.1084/jem.191.4.603
Celiac Disease Foundation. (2016). 20 Things You Might Not Know About Celiac Disease. Celiac Disease Foundation. https://celiac.org/about-the-foundation/featured-news/2016/08/20-things-you-might-not-know-about-celiac-disease/
Celiac Disease Foundation. (2023a). Symptoms of Celiac Disease. Celiac Disease Foundation. https://celiac.org/about-celiac-disease/symptoms-of-celiac-disease/
Celiac Disease Foundation. (2023b). What is Celiac Disease? Celiac Disease Foundation. https://celiac.org/about-celiac-disease/what-is-celiac-disease/
Dahal‐Koirala, S., Ciacchi, L., Petersen, J., Louise Fremgaard Risnes, Ralf Stefan Neumann, Christophersen, A., Knut E.A. Lundin, Hugh Harrington Reid, Qiao, S., Rossjohn, J., & Sollid, L. M. (2019). Discriminative T-cell receptor recognition of highly homologous HLA-DQ2–bound gluten epitopes. Journal of Biological Chemistry, 294(3), 941–952. https://doi.org/10.1074/jbc.ra118.005736
Hujoel, I. A., Jansson-Knodell, C., Hujoel, P. P., Margaux L.A. Hujoel, Rok Seon Choung, Murray, J. A., & Rubio–Tapia, A. (2020). Estimating the Impact of Verification Bias on Celiac Disease Testing. Journal of Clinical Gastroenterology, 55(4), 327–334. https://doi.org/10.1097/mcg.0000000000001361
Reynisson, B., Alvarez, B., Paul, S., Peters, B., & Nielsen, M. (2020). NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Research, 48(W1), W449–W454. https://doi.org/10.1093/nar/gkaa379
Singh, P., Arora, A., Strand, T. A., Leffler, D. A., Catassi, C., Green, P. H., Kelly, C. P., Ahuja, V., & Govind Makharia. (2018). Global Prevalence of Celiac Disease: Systematic Review and Meta-analysis. Clinical Gastroenterology and Hepatology, 16(6), 823-836.e2. https://doi.org/10.1016/j.cgh.2017.06.037
Wei, G., Helmerhorst, E. J., Darwish, G., Blumenkranz, G., & Schuppan, D. (2020). Gluten Degrading Enzymes for Treatment of Celiac Disease. Nutrients, 12(7), 2095–2095. https://doi.org/10.3390/nu12072095
Xia, J., Siegel, M., Bergseng, E., Sollid, L. M., & Khosla, C. (2006). Inhibition of HLA-DQ2-Mediated Antigen Presentation by Analogues of a High Affinity 33-Residue Peptide from α2-Gliadin. Journal of the American Chemical Society, 128(6), 1859–1867. https://doi.org/10.1021/ja056423o
Yin, L., & Stern, L. J. (2014). Measurement of Peptide Binding to MHC Class II Molecules by Fluorescence Polarization. Current Protocols in Immunology, 106(1). https://doi.org/10.1002/0471142735.im0510s106
Zhou, L., Kooy–Winkelaar, Y., Cordfunke, R. A., Dragan, I., Thompson, A., Drijfhout, J. W., Peter, & Koning, F. (2017). Abrogation of Immunogenic Properties of Gliadin Peptides through Transamidation by Microbial Transglutaminase Is Acyl-Acceptor Dependent. Journal of Agricultural and Food Chemistry, 65(34), 7542–7552. https://doi.org/10.1021/acs.jafc.7b02557
Feb Fair Poster